Dissecting regulatory non-coding variants in human disease
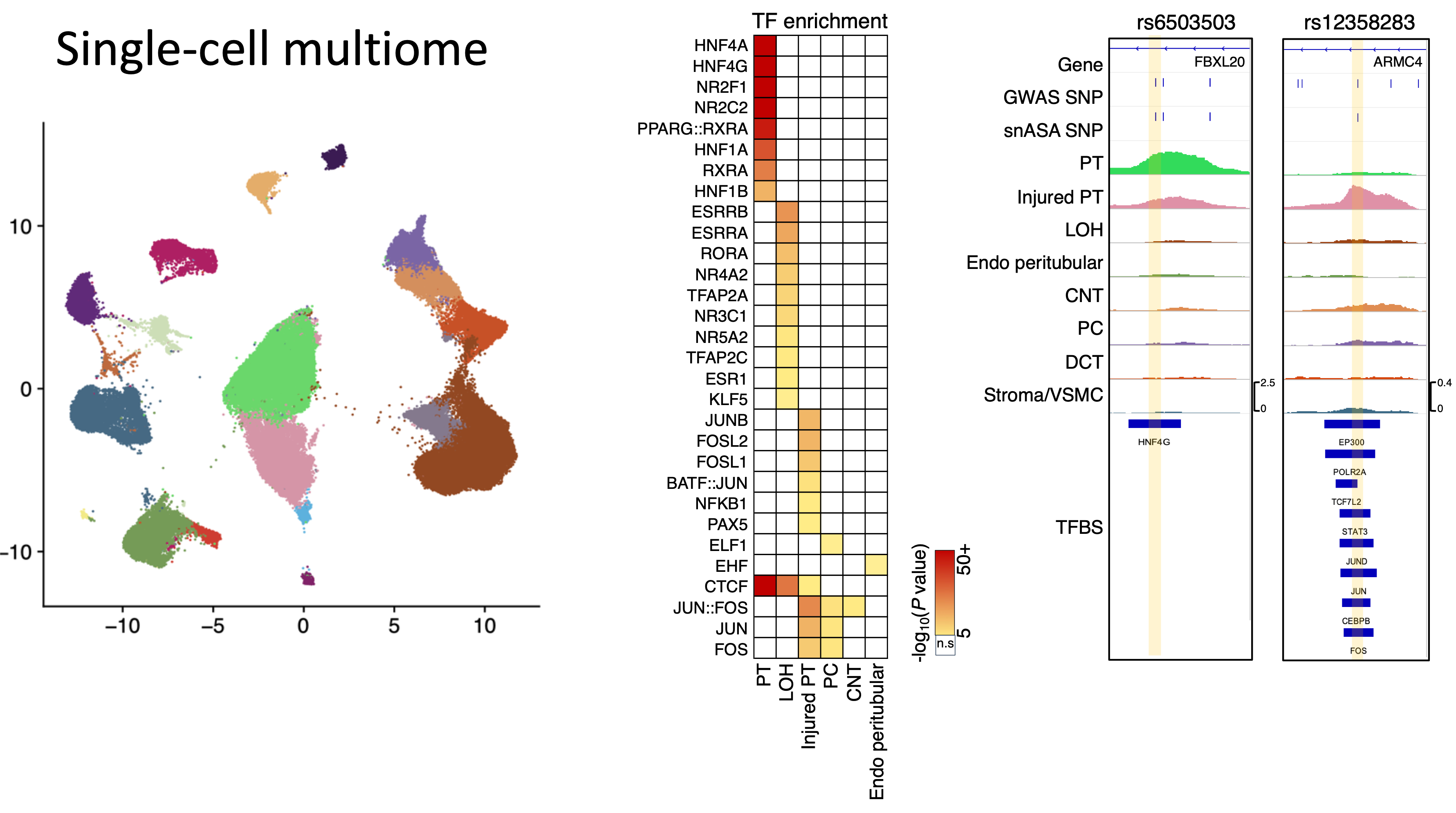
Genome-wide association studies (GWAS) have identified numerous DNA sequence variants associated with complex human diseases. However, over 90% of disease-associated variants reside in noncoding genome regions, and their functions in complex diseases remain largely unknown—a problem often referred to as the ‘variant-to-function’ problem. To dissect the regulatory function of noncoding DNA sequence variants, we perform large-scale genome-wide association studies (GWAS) and quantitative trait loci (QTL) studies for gene expression, DNA methylation and DNA accessibility. We discovered that epigenome (DNA methylation) explains a larger fraction of heritability than gene expression. Further, we found the allele-specific expression and open chromatin driven by GWAS variants associated with kidney disease. To further identify disease-causal genes, I proposed a Disease Genetic Scorecard and prioritized >600 kidney disease genes targeted by both coding variants and regulatory variants, including SLC47A1, whose causal role was defined in knockout mice model and human individuals carrying loss-of-function variants. (Liu et al., 2025 Science; Liu et al., 2022 Nature Genetics; Project page).
Epigenetic regulation in aging and age-related disease
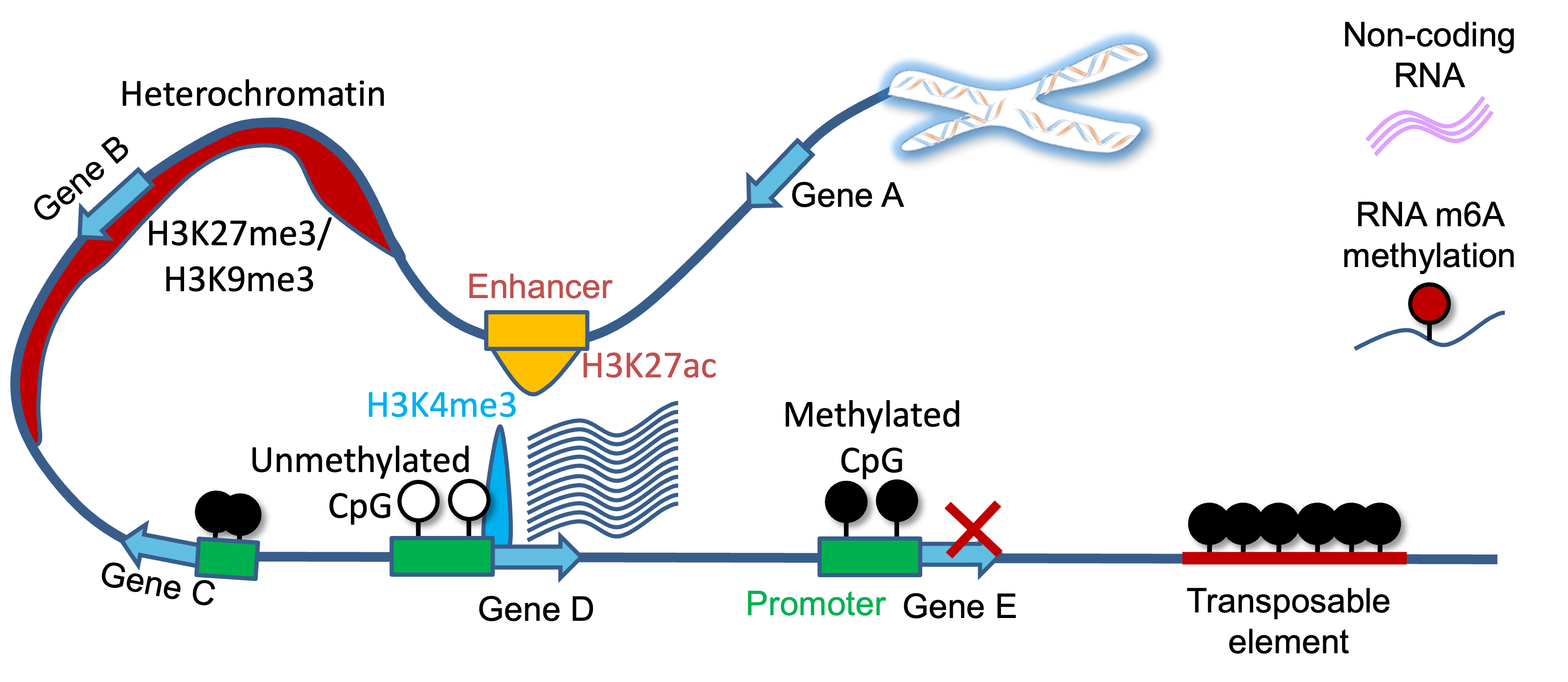
Epigenetic regulators play central roles in aging and age-related diseases. By applying our bioinformatic tools to large-scale epigenome data, we explore the epigenetic changes in aging and age-related chronic diseases. In previous studies, we have identified disease-critical epigenetic regulatory elements, including DNA methylation, histone modifications, super-enhancers, and long non-coding RNAs, and explored their roles in complex human diseases (e.g. kidney disease, diabetes, and cancer) (Liang, Liu et al. 2024 Nature Communications; Yan, Liu et al. 2024 Nature Communications; Xu, Liu et al. 2019 Cell Death & Disease; Xiong,…, Liu et al. 2017 NAR; Wei,…, Liu et al. 2016 NAR; Lv, Liu et al. 2013 NAR; Lv, Liu et al. 2012 NAR; Zhang, Liu et al. 2011 NAR; Zhang, Lv, Liu et al. 2010 NAR).
Identifying cellular origins of complex human disease
The causal cell type and regulatory mechanisms are poorly understood for complex diseases. To identify causal cell types, we developed a series of bioinformatic software, such as Open4Gene for linking enhancers to target genes from single cell multiome data (Liu et al., 2025 Science); SMART for de novo identifying tissue/cell-specific methylated regions from whole genome bisulfite sequencing data (Liu et al. 2016 NAR); QDMR for identifying differentially methylated regions from array-based methylation data (Zhang, Liu et al. 2011 NAR; Project page). Further, we generated single-nucleus transposase-accessible chromatin with sequencing (snATAC-seq), single-cell RNA sequencing (scRNA), and spatially resolved transcriptomics. The analysis illustrated the crucial roles of kidney cells (e.g., proximal tubule cells) and cell type-specific genes (e.g., SLC47A1) in kidney injury and fibrosis. (Liu et al., 2025 Science; Liu et al., 2022 Nature Genetics; Sheng,…,Liu et al., 2021 Nature Genetics; Miao,…, Liu et al. 2021, Nature Comms; Doke,…, Liu et al., 2021 JCI; Dhillion,…, Liu et al., Cell Metabolism; Abedini,…, Liu et al. 2022 BioRxiv).
Mapping epigenetic dynamics in mammalian development
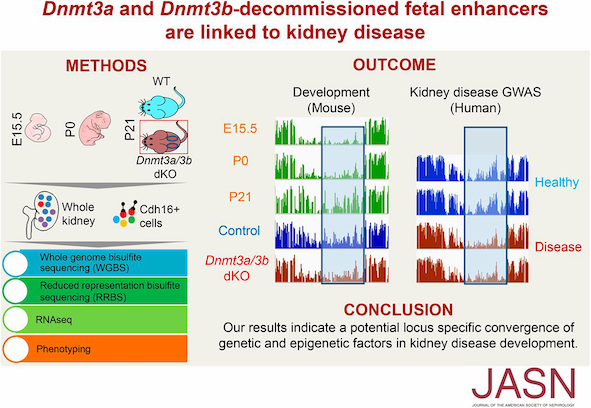
Epigenetic marking systems confer precise regulation of gene expression during cell development and differentiation. DNA methylation and histone modification undergo dynamics during mammalian development (Liu et al. 2014 Database; Liu et al. 2013 Sci. Rep.). Using knockout mice of Dnmt3a/b, we demonstrated essential roles of DNA methylation in decommissioned fetal enhancers linking to kidney disease (Guan, Liu et al. 2020 JASN). Using knockout mice of Tet2/3, we demonstrated critical roles of hydroxymethylation for nephron progenitor differentiation and nephron endowment (Liang,..., Liu et al. 2023 JASN). These studies highlighted locus-specific convergence of genetic, epigenetic, and developmental elements in disease development.